Introduction
There are very few posts on the internet that systematically talk about how to handle relatively large datasets in R. Especially, when the data size goes even larger, i.e. 32GB with 500k rows and 15k features, and consider the limitation of R language, loading them in a timely manner method is even challenging
There is almost zero information with a successful case to achieve the above situation.
However, in this post, I would introduce my method( after reading several posts and gave several tries) on I have loaded 32GB data, selected & filtered their columns, and exported them to new CSV data files in less than 30 minutes.
Procedure
Column Filter
It is ideal to create a column filter, either by column names or column indexes, to load only 2298 out of the entire 15000 column features into R.
For example, I only need feaures with names end with 0.0 in future analysis.
bd <- fread("/u/project/whoe_data.csv", header=TRUE,nrows = 2 )
bd_names <- colnames(bd)
bd_names1<-as.data.frame(strsplit(bd_names[-1],'-'))
bd_names2<-data.frame(t(bd_names1))
bd_names2<-bd_names2 %>% group_by(X1)%>% summarize(first = first(X2))
bd_names2$final_col<- paste(bd_names2$X1, bd_names2$first, sep="-")
final_col<-c('eid',bd_names2$final_col)
ind <- which(bd_names %in% final_col)
Load Data Rows By Chunks.
Laf is recommended in a few places, but when the data is very with, meaning when it has too many columns, fread() beats over LaF pretty much. And the complex nature of LaF using class method will also cause a lot of extra problems in practice, and takes more memories.
Also, though the fread() function from the library data.table appears in several posts on how powerful it is when reading
data files comparing to other method, it is nevered metioned by using for-loop and skip parameter, the data could also be load by chunks, for example 100000 rows in each loop, and the skip would help the pointer to go down by every 100000 and remember the locations.
This is my way of doing it:
data=NULL
for (i in 0:50){
data[[i+1]]=fread("/u/project/sriram/ukbiobank/33127/ukb39967.csv",nrow=10000,select=ind,skip =10000*i)
}
bd = data[[1]]
for (i in 1:50){
bd=rbind(bd,data[[i+1]],use.names=FALSE)
}
fwrite(bd,file="bd_ukb39967_3.csv")
Here we use loop and skip, nrows, to load data by chucks and then row-bind them in one large datasets bd.
Sometimes when there are too many loops, R will give warnings on memory limit. You may, for example, seperate one loop to three loops, excute each loop and then row-bind the three datasets into one by
rbind()orrbindlist().
In my case, it takes 20 mins for Jupyter R-kernel to complete
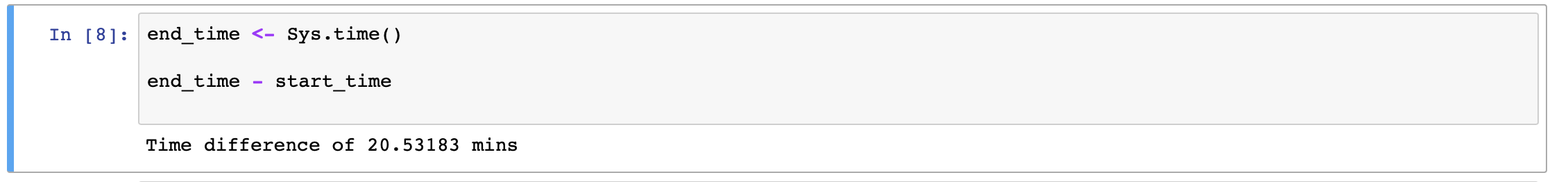
Testing and Conclusion
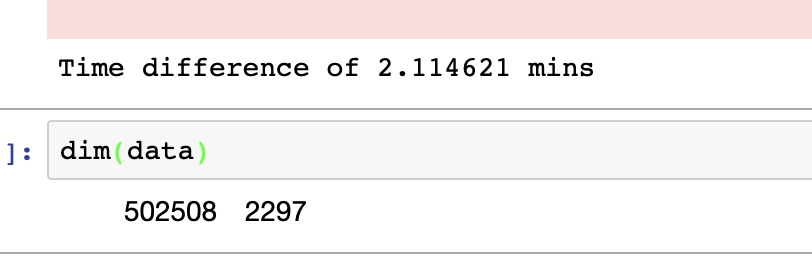
After saving my selected data columns with all thier rows in a new CSV files using fwrite() ,the data size goes from 32GB to 6 GB, and once I loaded it again, it takes about 1~2 mins.
The following shows the dimension and run-time for data loading.
The fread() is tested to be an outstanding way in R to load data exceeding 30GB, which is normally believed impossible to achieve in R in such short time!!!
Jiashu Miao edited on Feb, 5th, 2020.
Thanks for support and help from professor Eran Halperin and mentor Ph.D studetns at UCLA Bigdata Genomics Lab